- Research Article
- |
- Open Access
Negative association of prostate-derived ETS factor expression with estradiol induced molecules and estradiol levels in breast cancer; potential clinical implications
- Jianmin Wang;
- Department of Biostatistics and Bioinformatics, New York, USA
- Ashwani K Sood
- Immunology, Roswell Park Comprehensive Cancer Center, New York, USA

| Received | : | Dec 27, 2018 |
| Accepted | : | Jan 25, 2019 |
| Published Online | : | Jan 29, 2019 |
| Journal | : | Annals of Breast Cancer |
| Publisher | : | MedDocs Publishers LLC |
| Online edition | : | http://meddocsonline.org |
Cite this article: Wang J, Sood AK. Negative association of Prostate-derived ETS factor expression with estradiol induced molecules and estradiol levels in breast cancer; potential clinical implications. Ann Breast Cancer. 2019; 2(1): 1007.
Abstract
Background: Previous work from our and other laboratories showed that Prostate-Derived Ets Factor (PDEF) is a highly expressed oncogenic driver in breast cancer. However, the characteristics of PDEF expression in breast cancer and its relationship to estrogen signaling and estradiol levels remain poorly understood. To gain insights into these objectives, we analyzed the Cancer Genome Atlas (TCGA) breast cancer cohort consisting of 1095 breast tumors and the associated clinical and molecular data.
Methods: Specific Bioinformatics and Biostatistics methods outlined in the text were used.
Results: The results showed that 84% of breast tumors in this cohort cluster at high PDEF expression range. These included 96.4% of luminal (ER+ , PR+/-), 82.1% of Her2+ and 27.8% of triple negative tumors. Since virtually all ER+ , PR+/- tumors showed PDEF overexpression we tested whether PDEF expression was induced by estrogen signaling. Specifically, we determined PDEF expression in relation to other known estrogen induced molecules including PR, BRCA1, BRCA2 and NRIP1, in the ERhigh tumors from TCGA breast cancer cohort. The results showed a significant inverse correlation between PDEF expression and the expression of estrogen induced molecules. These findings are consistent with induction of PDEF expression during the loss of estrogen signaling, presumably to support the survival/growth of stressed epithelial tumor cells. To test this hypothesis, we compared PDEF expression in ER+ tumors arising from patients >55 years of age (as representative of postmenopausal status and low estradiol levels) with PDEF expression in tumors arising in patients <45 years of age (as representative of pre-menopausal status and high estradiol levels). This comparison showed significantly higher PDEF expression in tumors from >55 years age group compared to tumors from <45 years age group.
Conclusions: Our work shows negative association of PDEF expression with estradiol induced molecules and estradiol level. These findings have potential clinical implications. Specifically they suggest a role for PDEF in postmenopausal breast cancer risk and support targeting PDEF to minimize endocrine resistance in breast cancer.
Keywords: PDEF oncogene; SPDEF; Overexpression in breast tumors; Inverse correlation with estrogen signaling; Increased PDEF expression in tumors from older women
Introduction
Breast cancer is the most commonly diagnosed malignancy in females. Whereas the incidence of breast cancer remains high in Western countries, it is fast approaching alarming proportions in the developing countries of Asia and Africa as well [1-3]. The latter surge is attributed to increasing life expectancy and the adoption of western life styles by these populations. Over the years, a large effort has been devoted to developing preventive strategies to control the high burden of breast cancer worldwide. Specifically, chemoprevention based on the use of tamoxifen and the related inhibitors of signaling through Estrogen Receptor (ER) or inhibitors of estrogen synthesis have demonstrated the conceptual validity of this approach. About 50% of ER (estrogen receptor)+ breast cancers in high risk women could be prevented [4,5]. However, side effects including thrombosis, stroke, endometrial cancer and vasomotor symptoms, coupled with lack of biomarkers that predict response to these agents have limited the acceptance of this approach by most eligible women [6]. Evidently, there is an urgent need to understand additional molecular mechanisms that drive breast cancer with a view to developing novel approaches to breast cancer prevention.
Similarly, although the current treatments have significantly improved clinical outcomes, treatment-specific resistance remains a major barrier in breast cancer and contributes to metastatic progression in most patients. Again, novel molecular drivers of treatment resistance need to be identified to develop new approaches to minimize tumor progression.
We and others have previously reported that Prostate-Derived Ets Factor (PDEF), an Ets family transcription factor, is an oncogenic driver and a biomarker of poor prognosis in breast cancer (reviewed in 7). Specifically, transfection of PDEF into pre-malignant MCF-12A breast epithelial cell line led to increased tumorigenicity of PDEF expressing MCF-12A cells in immunodeficient mice [8]. Conversely, down regulation of PDEF by siRNA led to the loss of viability of BT-474 and SKBR3 breast tumor cell lines in vitro ([9]. Moreover, evidence for a pro-survival/growth role of PDEF in breast cancer was provided from the analysis of genes regulated by PDEF in the MCF-7 breast tumor cell line [10]. Specifically, gene ontology analysis of differentially regulated genes in PDEF-knockdown and control MCF-7 cells showed down regulation of cell cycle-related genes and upregulation of apoptosis-related genes in PDEF-knockdown cells [10].
Despite the significant oncogenic role of PDEF in breast cancer, its relationship to estrogen signaling in breast cancer remains poorly understood. A previous report using breast tumor cell lines concluded that PDEF expression was positively regulated by FoxA1 and ER [10]. In contrast, a genome wide study to identify genes that are regulated by estradiol reported that PDEF expression was suppressed by estradiol in MCF-7/BUS tumor cell line [11]. In the present study, we used bioinformatics approaches to analyze PDEF expression in primary tumors from TCGA (The Cancer Genome Atlas) breast cancer cohort and also conducted an analysis of PDEF expression in relation to estrogen induced molecules. Further, we compared PDEF expression in breast tumors from patients >55 years of age versus tumors from patients <45 years of age as representatives of postmenopausal and premenopausal status respectively. From this analysis we find that PDEF shows negative association with estradiol induced molecules and circulating estradiol levels. The significance of these findings to postmenopausal breast cancer risk is discussed. Also, these findings underscore the importance of PDEF as a novel target in breast cancer and support combining PDEF targeted approaches with endocrine approaches to minimize endocrine resistance in breast cancer.
Methods
Normalized gene expression data and clinical information are downloaded from TCGA data portal. Distribution of bimodal PDEF expression is fitted using mixtools R package with two normal distributions and all statistical tests are done using R statistical programming language. A p-value less than 0.05 is considered as statistical significant.
Results
Widespread PDEF overexpression in TCGA breast cancer cohort
Using a well-characterized anti-PDEF antibody, we previously reported PDEF protein expression in matched benign and tumor samples from breast cancer patients using immunohistochemistry [7]. In 8 of the 9 cases, PDEF was significantly overexpressed in the tumor tissue in comparison to the adjacent benign tissue. These results were also supported by our analysis of 131 primary breast tumors that showed 10-fold or higher expression of PDEF mRNA in 72% of these tumors compared to normal breast tissues [9].
To seek further confirmation of the high frequency of PDEF overexpression, we analyzed PDEF expression in the much larger TCGA dataset consisting of 1095 breast tumor samples. As shown in Figure 1, Panel A, a bimodal expression profile was observed with a high percentage (84%) of primary tumors clustering in the high PDEF expression range. The log 2 mean value for high PDEF expression group is 11.95 and for the low PDEF expression group is 6.88. On these bases, primary breast tumors comprising the high PDEF expression group showed mean PDEF overexpression of about 34-fold over the mean PDEF value for tumors that comprise the low PDEF expression group. We could not use normal breast tissue samples from TCGA cohort in this comparison since those tissues were selected for high epithelial content. Further analysis of PDEF expression within the breast tumor subtypes including luminal (ER+ , PR+/-) , Her2 positive, and triple negative subtypes showed that 96.4% of luminal, 82.1% of Her2 positive and 27.8% of triple negative tumors have high PDEF expression (data shown in Figure 1, Panel B). Together, these results showed widespread overexpression of PDEF in primary breast tumors form TCGA cohort that varied by the molecular subtype.
Figure 1: PDEF expression distribution in TCGA breast cancer cohort. Panel A, PDEF shows bimodal distribution with 84% tumors clustering at high expression range. Briefly, TCGA BRCA (breast cancer) RNASeq and clinical data were downloaded from TCGA data portal. Log2 transferred RSEM [12] normalized gene expression for level 3 RNASeq data are used for this analysis. Bimodal PDEF expression distribution was fitted into two Gaussian distributions with mean of 6.88 and 11.95 using mixtools package of R (http:// www.jstatsoft.org/article/view/v032i06) and plotted as solid curves. Panel B, frequency of PDEF overexpression in subtypes of breast cancers from TCGA cohort. Luminal subtype includes all tumors with ER+ and PR+/- status. ER and PR status is determined by IHC results. HER2 status is determined by IHC first, and for samples with equivocal HER2 IHC result and samples without HER2 IHC results, HER2 FISH results are used. The ER-, PR- samples that lacked information about the Her2 status were excluded from this analysis; as also were samples without ER and PR status information. Total tumor samples used in this analysis comprised 39 HER2, 825 Luminal and 158 TN/basal subtypes.
PDEF expression negatively correlates with estradiol induced molecules
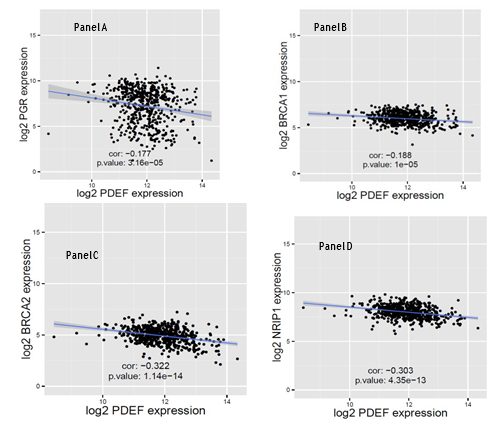
Since PDEF is overexpressed in 96.4% of luminal (ER+ , PR+/-) breast tumors, we tested whether PDEF expression is induced by estrogen signaling. Breast Cancer (BRCA) TCGA data, including clinical information and RSEM (RNA-Seq by Expectation Maximization) [12] normalized gene expression level, were downloaded from TCGA data portal. 1095 tumor samples with clinical and gene expression data were stratified into two groups (ERhigh and ERlow) according to the median ESR1 (estrogen receptor alpha gene) expression value. Using the ERhigh dataset, and considering PR (progesterone receptor) levels as a surrogate of estrogen signaling, (i.e., high PR levels denoting efficient estrogen signaling and low PR levels denoting poor estrogen signaling) we tested whether PDEF expression shows correlation with PR levels. As shown in Figure 2, Panel A, a significant inverse correlation between PDEF and PR expression was found (correlation coefficient: -0.177, p-value <0.0001). To obtain further evidence in support of this observation, PDEF expression was analyzed in relation to other molecules known to be induced by estradiol. These included BRCA1, BRCA2 and NRIP1 [11,13,14]. These markers were selected based on: i) their role as transcription factors or co-regulators of transcription with potential for significant impact on tumor cell biology and ii) they were previously identified as estrogen induced molecules in at least two of these three studies [11,13,14]. The results of this analysis are presented in Figure 2, panels B to D. Similar to PR, a significant inverse correlation between PDEF expression and the expression of BRCA1, BRCA2 and NRIP1 was observed. The relatively weaker negative correlation between PDEF and PR and PDEF and BRCA1 expression may be related to their loss of expression by mechanisms other than the loss of estrogen signaling. Specifically, mutations at the PGR locus may account for as much as 21% of ER+ tumors showing loss of PR expression [15], thereby undermining its negative correlation with PDEF that results solely from the loss of estrogen signaling. Similarly, the loss of BRCA1 expression due to mutations at the BRCA1 locus in 12% of ER+ tumors [16] may undermine its negative correlation with PDEF that results from the loss of estrogen signaling. Additionally, loss of BRCA1 expression may also occur due to methylation [17,18] and further contribute to the observed weaker negative correlation with PDEF. Nevertheless, the inverse correlation between PDEF versus BRCA2 and NRIP1 expression coupled with similar negative (although weaker) correlation of PDEF with PR and BRCA1 expression, strongly support the notion of negative association of PDEF expression with estradiol induced molecules in breast cancer. These results (presented in Figure 2, Panels A-D) to our knowledge provide the first evidence for negative association of PDEF expression with estradiol induced molecules in breast cancer.
Figure 2: Comparison of PDEF expression versus expression of estradiol induced molecules: Panels A to D, RNAseq data for 547 ERhigh breast tumors from TCGA were used for testing PDEF mRNA expression in relation to PR, BRCA1, BRCA2 and NRIP1 expression. Again, TCGA BRCA RNA-Seq and clinical data were downloaded from TCGA data portal. Log2 transferred RSEM normalized gene expression for level 3 RNA-Seq data were used for this analysis. Linear model was fitted to study the relationship between the gene pairs and Pearson correlation coefficients with p-values were carried out using R; p-value less than 0.05 is considered as statistical significant in the analysis. As shown, each of these molecules showed statistically significant inverse correlation with PDEF expression, although the strength of correlation varied. Specific explanations for weaker correlation for PR and BRCA1 are provided in the text.
Higher PDEF expression in tumors from postmenopausal age-group versus premenopausal age-group
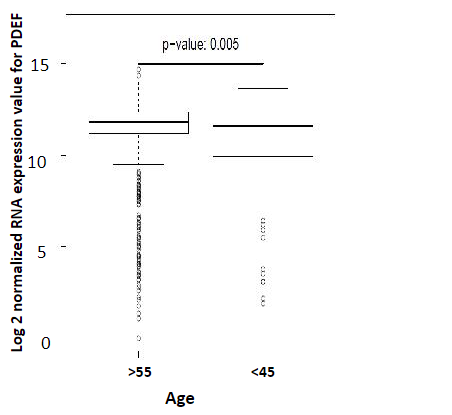
Since estradiol levels precipitously fall in postmenopausal women (to <10% of the premenopausal levels) [19], we tested whether PDEF expression increases in ER+ tumors arising in older women (due to loss of estrogen signaling) compared to tumors arising in younger women. Our analysis of the ERhigh subset of TCGA cohort showed that PDEF expression is indeed significantly higher in breast tumors from women >55 years of age (as representatives of postmenopausal status) compared to tumors from women <45years of age (as representatives of premenopausal status) The results are shown in Figure 3. These findings are consistent with the idea that loss of estradiol levels in postmenopausal years is conducive to increased PDEF expression in breast cancer
Figure 3: Comparison of PDEF expression in tumors from breast cancer patients >55 years of age versus <45 years of age: The ER- high subset of TCGA breast cancer cohort was analyzed for PDEF expression in tumors from these two age groups of patients, as representatives of postmenopausal and premenopausal status respectively.
Discussion
Few mutations at the PDEF gene locus were reported in the hundreds of breast cancers that have been sequenced to date. These results rule out a mutation-based mechanism contributing to PDEF overexpression in breast tumors. Based on FoxA1 binding to PDEF sequence and loss of PDEF expression following siRNA inhibition of FoxA1, FoxA1 was considered to positively regulate PDEF expression [10]. Moreover, this study showed that ER binds to PDEF sequence and based on this finding alone proposed that ER also positively regulates PDEF expression, even though siRNA inhibition of ER did not down regulate PDEF expression [10]. On the other hand, a genome wide study to identify genes that are regulated by estradiol reported the suppression of PDEF expression by estradiol in MCF-7/BUS tumor cell line [11]. Therefore the effect of estradiol on PDEF expression in breast cancer remained controversial. Our finding of a negative correlation between PDEF and estrogen induced molecules in primary breast tumors and its higher expression in breast tumors from older/posmenopausal patients show that PDEF expression is induced under conditions of loss of estrogen signaling, presumably to support the survival/growth of stressed epithelial tumor cells. These results are consistent with the notion that PDEF expression is suppressed by estradiol in breast cancer.
Significant downregulation of PDEF expression was reported at as low as 10 pM concentration of estradiol, with continuing increased downregulation (to <30% of control levels) occurring at higher concentrations (up to 100 pM) of estradiol [11]. It is noteworthy that 100 pM (27.2 pg/ml) serum estradiol concentration may occur in vivo primarily in pre-menopausal women [19]. Hence strong suppression of PDEF expression by estradiol may occur mostly in premenopausal women, whereas most postmenopausal women exhibiting weaker PDEF suppression. Additionally, besides the loss of estrogen signaling, loss of GATA3 expression also occurs in breast tumors from postmenopausal women [20], presumably partly due to loss of estrogen signaling since ER induces GATA3 expression [21], and/or due to frequent structural mutations in GATA3 gene in as many as 17% of ER+ breast tumors [22]. Since GATA3 is also a negative regulator of PDEF expression [10], increased loss of GATA3 in postmenopausal tumors could provide another mechanism for PDEF induction in tumors arising in postmenopausal years. These findings have implications for a role for PDEF in increased risk of breast cancer with advancing age in postmenopausal women (see below).
Prolonged exposure to estrogens has long been recognized as a major risk factor for breast cancer development. However, estrogen levels dip drastically following menopause [19], yet breast cancer incidence continues to rise in postmenopausal years as about 80% of breast cancers may occur in women >55 years of age. Mutational activation of driver genes appears inadequate to explain the high breast cancer incidence especially since ER+ breast cancers have low number of nonsynonymous mutations [23]. Evidently other mechanisms need to be invoked that contribute to the high incidence of breast cancer in postmenopausal years, with continuing increased rate of breast cancer risk with advancing age well into the 70s. To that end, a recent genome wide mapping of cancer dependencies showed that in >80% of tumor models, the top cancer dependency biomarkers were derived from changes in the expression of specific driver genes rather than the mutation-based functional alteration of these genes [24]. In that study, SPDEF (another name for PDEF) was identified as an important lineage restricted driver of breast cancer [24]. Based on this understanding and the widespread PDEF overexpression in ER+ breast tumors we believe that ER+ breast tumors in postmenopausal women are dependent on both estrogen signaling and on PDEF expression for survival and growth, thereby PDEF conferring a continuing increased risk of breast cancer during decreasing estradiol levels coupled with loss of GATA3 expression in tumors arising at increasing age in postmenopausal years.
Our finding of an inverse correlation between PDEF and estrogen induced molecules in ER+ breast tumors is both novel and interesting especially since 96.4 % of ER+ breast tumors show overexpression of PDEF. The increased PDEF expression presumably compensates for the reduced estrogen signaling, especially in tumors from postmenopausal women, by supporting the survival of stressed epithelial tumor cells. Together these two oncogenic pathways may drive the development and progression of ER+ breast tumors in postmenopausal women. This may be why targeting the estrogen signaling axis only prevents about 50% of ER+ breast tumors in chemoprevention trials [4,5] and why endocrine therapy engenders resistance in the treatment of ER+ breast tumors. This idea is also supported by the observed poor overall survival of patients with PDEF overexpressing ER+ breast tumors [8,10]. Together, these findings provide rationale for dual targeting of ER+ breast tumors with endocrine therapy and PDEF targeted approaches. Similarly, a high proportion (82.1%) of Her2+ breast tumors overexpress PDEF and these also appear to be partly driven by PDEF. This idea is supported by our findings that down regulation of PDEF expression by siRNA leads to the loss of the viability of Her2 overexpressing BT-474 and SKBR3 human breast tumor cell lines in vitro [9], suggesting that these cell lines are also dependent on PDEF besides Her2 for growth and survival. Hence, targeting PDEF in conjunction with Her2 should be beneficial in minimizing resistance to Her2 targeted approaches. Likewise, overexpression of PDEF in 27.8% of triple negative breast tumors supports the development of PDEF targeted approaches in conjunction with existing treatments for these tumors, for which targeted therapies do not exist at present. Overall there is a compelling rationale for developing PDEF-targeted drugs and vaccines to combine with existing approaches for prevention and treatment of breast cancer.
Acknowledgement
This work was supported by grants from Breast Cancer Coalition of Rochester (#62-3218-01) and Roswell Park Alliance Foundation (#62-2562-01). The core resources used in this research were funded by the Cancer Center Support Grant P30CA16056 to Roswell Park Cancer Institute.
Contributions
JW, Hypothesis development, data collection, data interpretation, literature search and writing and A.K.S, Hypothesis development, data collection, data interpretation, literature search and writing.
References
- Sung H, Rosenberg PS, Chen WQ, Hartman M, Lim WY, et al. Female breast cancer incidence among Asian and Western populations: more similar than expected. J Natl Cancer Inst. 2015; 107:1-7.
- Goss PE, Strasser-Weippl K, Lee-Bychkovsky BL, Fan L, Li J, et al. Challenges to effective cancer control in China, India, and Russia. Lancet Oncol. 2014; 15: 489-538.
- Akarolo-Anthony SN, Ogundiran TO, Adebamowo CA. Emerging breast cancer epidemic: evidence from Africa. Breast Cancer Res. 2010; 12: S8.
- Vogel VG, Costantino JP, Wickerham DL, Cronin WM, Cecchini RS, et al. National Surgical Adjuvant Breast and Bowel Project. Update of the National Surgical Adjuvant Breast and Bowel Project Study of Tamoxifen and Raloxifene (STAR) P-2 Trial: Preventing breast cancer. Cancer Prev Res (Phila). 2010; 3: 696-706.
- Cuzick J, Sestak I, Forbes JF, Dowsett M, Knox J, et al. IBIS-II investigators. Anastrozole for prevention of breast cancer in highrisk postmenopausal women (IBIS-II): an international, doubleblind, randomised placebo-controlled trial. Lancet. 2014; 383: 1041-1048.
- Fagerlin A, Dillard AJ, Smith DM, Zikmund-Fisher BJ, Pitsch R, et al. Women’s interest in taking tamoxifen and raloxifene for breast cancer prevention: response to a tailored decision aid. Breast Cancer Res Treat. 2011; 127: 681-688.
- Sood AK, Geradts J, Young J. Prostate-derived Ets factor, an oncogenic driver in breast cancer. Tumor Biol. 2017; 39: 1-6.
- Sood AK, Wang J, Mhawech-Fauceglia P, Jana B, Liang P, et al. Sam-pointed domain containing Ets transcription factor in luminal breast cancer pathogenesis. Cancer Epidemiol Biomar & Pev. 2009; 18: 1899-1903.
- Mukhopadhyay A, Khoury T, Stein L, Shrikant P, Sood AK. Prostate derived Ets transcription factor and Carcinoembryonic antigen related cell adhesion molecule 6 constitute a highly active oncogenic axis in breast cancer. Oncotarget. 2013; 4: 610-621.
- Buchwalter G, Hickey MM, Cromer A, Selfors LM, Gunawardane RN, et al. PDEF promotes luminal differentiation and acts as a survival factor for ER-positive breast cancer cells. Cancer Cell. 2013; 23: 753-767.
- Coser KR, Chesnes J, Hur J, Ray S, Isselbacher KJ, et al. Global analysis of ligand sensitivity of estrogen inducible and suppressible genes in MCF7/BUS breast cancer cells by DNA microarray. Proc Natl Acad Sci U S A. 2003; 100: 13994-13999.
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12: 323-338.
- Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, et al. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003; 144: 4562-4574.
- Lin CY, Ström A, Vega VB, Kong SL, Yeo AL, et al. Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol. 2004; 5: R66.
- Mohammed H, Russell IA, Stark R, Rueda OM, Hickey TE, et al. Progesterone receptor modulates ERα action in breast cancer. Nature. 2015; 523: 313-317.
- Rhiem K, Todt U, Wappenschmidt B, Klein A, Wardelmann E, et al. Sporadic breast carcinomas with somatic BRCA1 gene deletions share genotype/phenotype features with familial breast carcinomas. Anticancer Res. 2010; 30: 3445-3449.
- Otani Y, Miyake T, Kagara N, Shimoda M, Naoi Y, et al. BRCA1 promoter methylation of normal breast epithelial cells as a possible precursor for BRCA1-methylated breast cancer. Cancer Sci. 2014; 105: 1369-76.
- Hosny MM, Sabek NA, El-Abaseri TB, Hassan FM, Farrag SH. Promoter Methylation Status of Breast Cancer Susceptibility Gene 1 and 17 Beta Hydroxysteroid Dehydrogenase Type 1 Gene in Sporadic Breast Cancer Patients. Int J Breast Cancer. 2016 : 105: 1369-1376.
- Rothman MS, Carlson NE, Xu M, Wang C, Swerdloff R, et al. Reexamination of testosterone, dihydrotestosterone, estradiol and estrone levels across the menstrual cycle and in postmenopausal women measured by liquid chromatography-tandem mass spectrometry. Steroids. 2011; 76 : 177-182.
- Hosoda M, Yamamoto M, Nakano K, Hatanaka KC, Takakuwa E, et al. Differential expression of progesterone receptor, FOXA1, GATA3, and p53 between pre- and postmenopausal women with estrogen receptor-positive breast cancer. Breast Cancer Res Treat. 2014; 144: 249-261.
- Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, et al. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 2007; 67: 6477- 83.
- Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012; 486: 400-404.
- Budczies J, Bockmayr M, Denkert C, Klauschen F, Lennerz JK, et al. Classical pathology and mutational load of breast cancer-integration of two worlds. J Pathol Clin Res. 2015; 1: 225-238.
- Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, et al. Defining a Cancer Dependency Map. Cell. 2017; 170: 564- 576.
MedDocs Publishers
We always work towards offering the best to you. For any queries, please feel free to get in touch with us. Also you may post your valuable feedback after reading our journals, ebooks and after visiting our conferences.


![Figure 1: PDEF expression distribution in TCGA breast cancer
cohort. Panel A, PDEF shows bimodal distribution with 84% tumors
clustering at high expression range. Briefly, TCGA BRCA (breast cancer) RNASeq and clinical data were downloaded from TCGA data
portal. Log2 transferred RSEM [12] normalized gene expression
for level 3 RNASeq data are used for this analysis. Bimodal PDEF
expression distribution was fitted into two Gaussian distributions
with mean of 6.88 and 11.95 using mixtools package of R (http://
www.jstatsoft.org/article/view/v032i06) and plotted as solid
curves. Panel B, frequency of PDEF overexpression in subtypes of
breast cancers from TCGA cohort. Luminal subtype includes all tumors with ER+ and PR+/- status. ER and PR status is determined by
IHC results. HER2 status is determined by IHC first, and for samples
with equivocal HER2 IHC result and samples without HER2 IHC results, HER2 FISH results are used. The ER-, PR- samples that lacked
information about the Her2 status were excluded from this analysis; as also were samples without ER and PR status information.
Total tumor samples used in this analysis comprised 39 HER2, 825
Luminal and 158 TN/basal subtypes. ...](figures/PDEF-expression-distribution-in-TCGA.jpg)