- Research Article
- |
- Open Access
Manual vs Digital Scoring of Ki67 in Breast Cancer using an Automated Image Analysis System: An Interobserver Variability Study
- Ahsan Siddiqi, MD;
- Department of Pathology and Laboratory Medicine, Baylor University, Houston, Texas, USA.
- Helena Hanna;
- student at The Bolles School, Jacksonville, Florida, USA.
- Xochiquetzal J Geiger, MD;
- Department of Laboratory Medicine and Pathology, Mayo Clinic, Jacksonville, Florida, USA.
- Aziza Nassar, MD, MPH, MBA*
- Department of Laboratory Medicine and Pathology, Mayo Clinic, Jacksonville, Florida, USA.

| Received | : | Feb 19, 2021 |
| Accepted | : | Mar 15, 2021 |
| Published Online | : | Mar 18, 2021 |
| Journal | : | Annals of Breast Cancer |
| Publisher | : | MedDocs Publishers LLC |
| Online edition | : | http://meddocsonline.org |
Cite this article: Siddiqi A, Hanna H, Geiger XJ, Nassar A. Manual vs Digital Scoring of Ki67 in Breast Cancer using an Automated Image Analysis System: An Interobserver Variability Study. Ann Breast Cancer. 2021; 4(1): 1017.
Keywords: Breast cancer; Digital imaging; Image analysis; Ki67; Manual assessment.
Abbreviations: BC: breast cancer; DIA: digital image analysis; ER: estrogen receptor; ICC: intraclass correlation coefficient; LI: labeling index; PR: progesterone receptor; VA: visual assessment.
Keypoints
There is wide variability among pathologists when using visual assessment for Ki67 (MIB-1) in breast cancer.
Digital image analysis eliminates interobserver variability of Ki67 assessment in breast cancer, among pathologists.
Evaluating Ki67 using the hot spot method with automated image analysis appears to be superior to manual visual assessment.
Abstract
Objectives: We sought to validate the use of Digital Image Analysis (DIA) with the Aperio Digital Scanner (Leica Biosystems) for assessment of Ki67 proliferation Labeling Index (LI) in patients with Breast Cancer (BC) and compare the results with the manual method of Visual Assessment (VA).
Methods: We retrospectively identified 87 patients with BC and retrieved paraffin-embedded, whole-tissue sections for Ki67 immunostaining (monoclonal MIB-1 clone). Two pathologists independently reviewed and annotated Ki67 LI using VA. The sections were also subjected to DIA Ki67 quantitation for hot spots using the Aperio system. Bland-Altman analysis was used to evaluate agreement between VA and DIA.
Results: There was wide variation in Ki67 LI score by VA between the 2 pathologists, with mean Ki67 scores of 9.2% and 6.2%. The DIA reported a mean Ki67 score of 9.4%. By Bland-Altman analysis, DIA showed a mean difference of 0.1 vs pathologist 1 and 3.2 vs pathologist 2. Scores were significantly different between the 2 pathologists and between DIA and pathologist 2 (both P<.001). Scores demonstrated excellent agreement between pathologist 1 and DIA (P=.84).
Conclusions: Our study validates the use of DIA for providing more reliable Ki67 LI assessment to mitigate interobserver variability among pathologists.
Introduction
Ki67 is a proliferation biomarker that is expressed in all cells during all growth cell cycles except for the resting phase [1]. In patients with Breast Cancer (BC), Ki67 expression has been found to correlate with disease-free and overall survival [1,2], to predict response to chemotherapy in the neoadjuvant setting [3,4], and to act as a marker to predict response to neoadjuvant endocrine therapy [5]. Furthermore, it is a better tool than the mitotic activity index to assess prognosis for risk recurrence in patients who are pretreated with endocrine therapy.
Recent data suggest that a Ki67 proliferation Labeling Index (LI) higher than 10% to 14% defines a high-risk group in terms of prognosis in BC [5,6]. In the ACOSOG Z1031 trial [7], patients were triaged to standard chemotherapy when their tumors exhibited a Ki67 LI greater than 10% 2 to 4 weeks after starting aromatase inhibitors. The POETIC* trial [8] showed that Ki67 LI at baseline and 2 weeks after aromatase inhibitor therapy can predict patients who are most likely to have increased risk of recurrence and hence require additional chemotherapy. Recurrence risk was lower (4.5%) for patients with low Ki67 LI (<10%) at baseline and 2 weeks, but risk was higher (19.6%) for patients with a high Ki67 LI (≥10%) at baseline and 2 weeks [8].
Assessment of LI using Ki67 (MIB-1), however, can be difficult and has been inconsistent. The International Ki67 in Breast Cancer Working Group recommended counting a minimum of 500 tumor cells and an ideal of at least 1,000 cells when evaluating Ki67 in patients with BC [9]. The main issue with assessment of Ki67 LI is the lack of reproducibility, with its inherent wide interlaboratory and intralaboratory variability, especially for Visual Assessment (VA). Several groups have evaluated the utility of automated image analysis for quantification of Ki67 expression to eliminate the variability between and within laboratories and among pathologists when using VA [10-12].
In the current study, we aimed to evaluate VA and automated assessment of Ki67 LI between 2 pathologists and an automated scanner in BC. Our primary hypothesis was that automated assessment of Ki67 LI might improve interpathologist variability for scoring Ki67 as a proliferation biomarker.
Materials and methods
This study was approved by our Mayo Clinic institutional review board, and patients were consented under the IRB# 15-006965. Using our pathology database, we retrospectively searched for the records of patients seen from January 1, 2012, through December 31, 2016, who had a BC diagnosis that was categorized as either luminal subtype A or B and who had archived tissue available for retrieval. For all patients who met our search criteria and were randomly selected, demographic and clinical information was obtained from the electronic health record, and whole-tissue sections of formalin-fixed, paraffin-embedded blocks were retrieved.
We then performed immunohistochemical staining of the whole-tissue sections with Ki67 immunostain (monoclonal MIB-1 clone; Agilent) and the Ventana’s Ultraview detection system. MIB-1 monoclonal antibody was diluted 1:20 using the ultraView Universal DAB Detection Kit (Ventana Medical Systems, Inc). After pretreatment with high-pH ULTRA Cell Conditioning Solution (Ventana) for 30 minutes, slides were incubated with MIB-1 antibody for 32 minutes at 37 °C.
The slides’ images are organized using E-slide manager (2006-2018), Leica Biosystems (Version 12.4.2.5010). The images are analyzed using Digital Image Analysis via Aperio Image Scope software version 12.4.2.5010 (Leica Biosystems Pathology Imaging 2003-2018). A technologist selects “Hot” regions of the most positive areas of the tumor. A minimum of 2000 cells are selected. The results are reported out as the percent positive out of total number of cells. The slides and images are reviewed by the pathologist who confirms final interpretation.
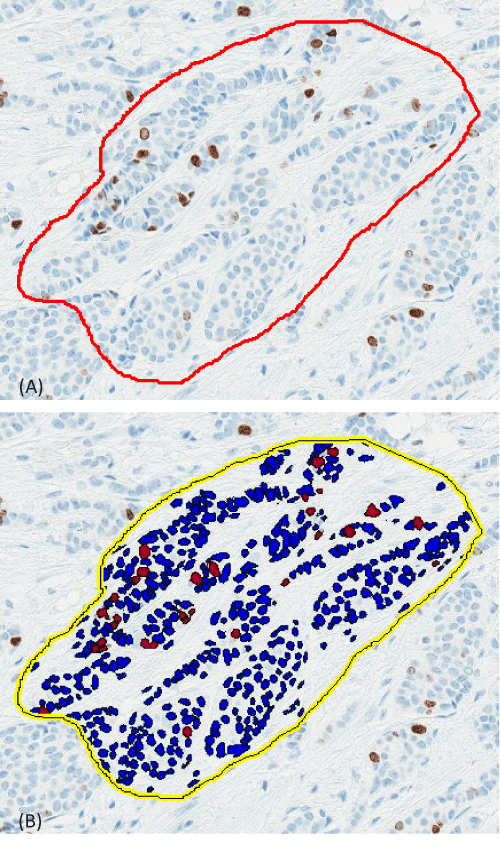
Two pathologists, designated pathologist 1 and pathologist 2, independently and blindly reviewed the slides and annotated the Ki67 results, using average VA, according to their expertise. The results are reported as the percentage of positively stained tumor nuclei over the total tumor nuclei, termed LI. Separately, 2 histotechnologists performed Digital Image Analysis (DIA) quantitation of the slides on an Aperio Digital Scanner (Leica Biosystems). One histotechnologist scored part of the slides and the other the rest of the slides and together they scored 100% of slides. Quantitation of MIB-1 staining by DIA on the Aperio system in breast tumors has been validated in our laboratory [13,14]. The results of DIA focused on hot spots in the tumor periphery, which have shown the best predictive value [15]. The histotechnologists trained the Aperio system to omit lymphocytes and include ony tumor cells when making the digital annotation. Furthermore, the technologists used the pen tool when necessary to eliminate the areas with stromal cells as much as possible when there is significant number of stromal cells. They also tried to circle areas with a large amount of tumor cells and insignificant amount of stromal cells. If there is a disagreement between the pathologists’ read and the DIA, then a consensus agreement was reached as which method was deemed the most accurate. Figure 1 illustrates the accuracy of the digital scanner for determining positive vs negative staining. The only concern is the location selected for counting of cells. The results are confirmed by the pathologist after the tissue is analyzed, which allows the pathologist to designate another area to be analyzed, if necessary, before releasing the result.
Statistical analysis
Descriptive statistics were used for the demographic and clinical data. Bland-Altman plots [16] were used to evaluate the agreement in Ki67 results between the VA and the DIA. Paired t tests were used to assess for significant differences between the 2 independent reviewers and between each reviewer and DIA. Analysis was completed using SAS version 9.4 (SAS Institute Inc), and P<.05 was considered statistically significant.
Results
We identified 87 women with a BC diagnosis (luminal subtype A or B) who were included in the study. The average age of the cohort was 66.8 years (range, 47-89 years). The tumor subtypes were invasive ductal carcinoma (71; 82%), invasive lobular carcinoma (13; 15%), and mixed carcinoma (3; 3%). Tumor grades using the Nottingham grading system were grade 1 for 29 patients (33%), grade 2 for 49 patients (56%), and grade 3 for 9 patients (10%). The average size of the tumor was 1.5 cm (range, 0.7-5.0 cm). All 87 cases (100%) were Estrogen Receptor (ER) positive, and 81 (93%) were Progesterone Receptor (PR) positive. ERBB2 (formerly HER2/neu) was positive in 1 case (1%), negative in 84 cases (97%), and equivocal in 2 cases (2%). Treatment included chemotherapy for 24 patients (28%) and radiotherapy for 62 patients (71%); all 87 (100%) were treated with endocrine therapy. Disease recurrence was noted in only 3 patients (3%), who had an average follow-up of 128 months (range, 12-192 months).
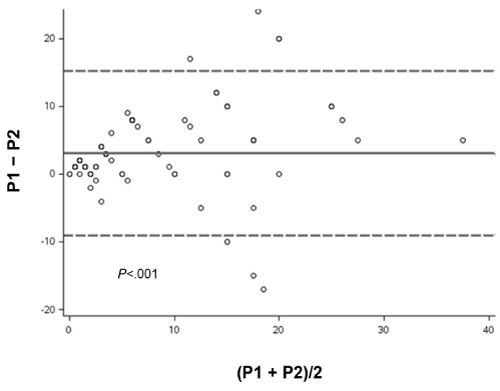
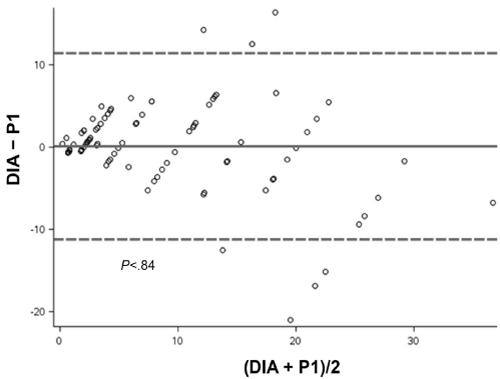
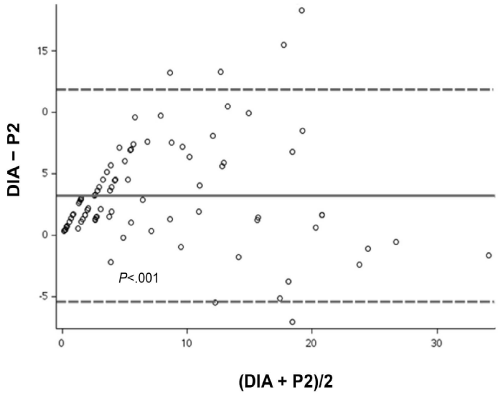
In the VA by the 2 pathologists, pathologist 1 reported a mean Ki67 LI score of 9.2%, and pathologist 2 reported a mean score of 6.2% (Table). Bland-Altman analysis of the difference between the VA Ki67 score of both pathologists showed a significant difference (mean, 3.1%; P<.001) (Figure 2). The DIA reported a mean Ki67 score of 9.4%. Bland-Altman analysis showed no significant difference in scores between DIA and pathologist 1 (mean, 0.1%; P=.84) (Figure 3). In contrast, analysis of scores for DIA vs pathologist 2 showed a significant difference (mean, 3.2%; P<.001) (Figure 4).
The Ki67 LI scores by DIA were then divided into 3 groups: low, <14% (n=64); intermediate, 14%-20% (n=12); and high, >20% (n=11). The median (Range) difference between the VA measures of the 2 pathologists for the same groups were 1% (–4% to 24%) for the low group, 5% (–10% to 12%) for the intermediate group, and 5% (–17% to 20%) for the high group. Across the 3 groups, the variability was lower for the low group, but the difference was not statistically significant (Kruskal-Wallis P=.33). After careful review of cases that were discrepant between the 2 pathologists and with DIA, DIA was deemed to be more accurate.
Figure 1: Immunostained Slides Showing Ki67 Digital Scoring. Slides were immunostained for Ki67 (monoclonal MIB-1 clone; Agilent), and digital scoring was performed on the Aperio Digital Scanner (Leica Biosystems). The area analyzed is outlined in red, with blue demonstrating negative cells and brown demonstrating positive cells (A; without mask); and this is the image with segmentation utlined in yellow with dark blue cells showing negative tumor cells and bright red showing positive tumor cells with Ki67 (B; with mask).
Figure 2: Bland-Altman Plot. Variation in mean Ki67 scores by visual assessment between pathologist 1 (P1) and pathologist 2 (P2), analyzed by paired t test. Solid line indicates mean difference; dashed lines indicate 95% CI.
Figure 3: Bland-Altman Plot. Variation in mean Ki67 scores between Digital Image Analysis (DIA) and visual assessment by pathologist 1 (P1), analyzed by paired t test. Solid line indicates mean difference; dashed lines indicate 95% CI.
Figure 4: Bland-Altman Plot. Variation in mean Ki67 scores between Digital Image Analysis (DIA) and visual assessment by pathologist 2 (P2), analyzed by paired t test. Solid line indicates mean difference; dashed lines indicate 95% CI.
Discussion
In the current study, the analysis of Ki67 staining with VA by pathologist 1 and DIA using the Aperio system demonstrated excellent agreement. When analysis was performed manually, the results were not always consistent between reviewers, especially when there was heterogeneity in the expression of Ki67 within the tumor. We believe that use of DIA will provide the patient with more consistent results. Our results clearly demonstrate how digital scoring can be used to mitigate the interobserver variability seen with manual analysis.
Increased Ki67 expression is a predictor of increased response to neoadjuvant chemotherapy [17]. It is also a surrogate marker of achieving pathologic complete response because highly mitotically active tumors respond well to neoadjuvant chemotherapy, and with a threshold of 28% LI (area under the receiver operating characteristic curve, 0.89; 95% CI, 0.75-0.96) has been shown to predict tumor regression after neoadjuvant chemotherapy [4]. In addition, an increased Ki67 LI is associated with worse prognosis and has been shown to be an independent prognostic marker for decreased recurrence-free and overall survival [2,9]. Furthermore, it was found to be a predictive marker of endocrine resistance in the neoadjuvant setting which necessitates more aggressive treatment [7]. In addition, high Ki67 LI has been significantly associated with larger tumor size, higher tumor grade, more nuclear pleomorphism, and increased mitotic scores [18], as well as with higher tumor stages and higher nodal status [2]. Interestingly, invasive ductal carcinomas tend to have a high level of Ki67 expression (LI, 22%), whereas invasive lobular cancers tend to have lower mean Ki67 expression (LI, 13%) [2]. One study found little variability in Ki67 expression in BC, with median Ki67 LI values of 15.0% to 17.5% over 4 years in ER/PR-positive BC, 55.0% to 60.0% for triple-negative cases, and 30.0% to 32.5% for ERBB2-positive cases [19].
Ki67 clearly has prognostic importance, but because of intratumoral heterogeneity and low analytical validity for Ki67 LI, it is difficult to standardize Ki67 LI assessment across laboratories due to lack of reproducibility [1]. One study showed that a predefined value of 15% in at least 500 to 1,000 counted cells is needed to reach an acceptable error rate [1].
Many different cutoff values for Ki67 have also been described for different prognostic settings. A cutoff Ki67 LI value of 20% was found to be the best marker to designate high-risk patients with luminal-type BCs [20]. That study showed that Ki67 LI, together with tumor size and lymph node status, could identify patients with ER-positive BC who need combined chemotherapy and hormonal therapy because of poor prognosis [20]. A Ki67 LI cutoff of 20% or greater using tissue microarray was found to be the best prognostic cutoff, particularly for ERBB2-positive and triple-negative BC. In 2011, the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer recommended an alternative Ki67 cutoff point of 14% to separate ER-positive tumors into luminal A (<14%) and luminal B (≥14%) subtypes [6]. A cutoff point of 20% in BC when assessing Ki67 LI on tissue microarrays appears to be optimal for both concordance with whole-tissue section values and predicting patient outcome [18].
Some studies have shown excellent agreement between VA and automated DIA of Ki67 LI in BC [12]. However, the method of scoring seems to be important, such as average vs hot spot scoring. The average and hot spot scoring methods demonstrated perfect concordance between VA and DIA for Ki67 LI in one study, with only slightly better agreement for the average scoring method (Intraclass Correlation Coefficient [ICC], 0.974; 95% CI, 0.964-0.981; P<.001) than the hot spot method (ICC, 0.957; 95% CI, 0.941-0.968; P<.001) [12]. These findings are similar to those of other groups [21]. In another study, the counting system (ICC, 0.66; 95% CI, 0.52-0.78) had better concordance among pathologists than the scoring (visual estimate) system (ICC, 0.57; 95% CI, 0.42-0.72), especially when the assessed field was preselected [22].
There is excellent agreement both within and between DIA platforms. Intraplatform reproducibility has been excellent for all investigated DIA platforms (ICC, 0.972-0.992) and among operators (ICC, 0.962-0.995) [23]. In one study, DIA reduced not only intraobserver but also interobserver variability [24]. The interobserver variability of Ki67 LI for direct counting and categorical estimation was relatively high. That study recommended performing Ki67 LI measurement using direct counting rather than rough categorical estimation because of less interobserver variability in the former [24]. Tumors that exhibited hot spots generally showed greater interobserver variability than those without hot spots. In addition, when the areas of Ki67 measurement were restricted to the tissue microarray platform, the interobserver variability decreased, even when the tumors displayed hot spots. Therefore, that study and others suggested that a specific area should be selected when evaluating Ki67 LI, particularly the periphery of the tumor or hot spots [24,25].
One study proposed using a stepwise counting strategy that specifically evaluates small, highly proliferative hot spots [25]. Kwon et al [11] confirmed that VA and automated DIA are highly correlated (ICC, 0.982), albeit after an expert confirms the results of the automated DIA. They also noted that differences between VA and DIA were due to multiple factors, including tumor heterogeneity [25], VA interpretation errors, misidentification of tumor cells, poor immunostaining or slide quality, and estimation of nontumoral cells [11,24].
Scoring concordance, both between methods and between observers, also seems to depend on the Ki67 LI. As with previous studies [21,26], Kwon et al [11] noted that the intermediate Ki67 LI group (10%-20%) showed relatively weak concordance between VA and DIA. Regarding interobserver variability, a multicenter collaboration revealed that observers scoring Ki67 LI show excellent to perfect concordance on cases that are either much lower or higher than the intermediate range [27]. Furthermore, lower rates of agreement between observers are noted mainly for intermediate Ki67 LI (>10%-15%) in BC [28]. In our current study, although interobserver variability was higher in the intermediate and high Ki67 categories, the differences were not statistically significant.
Some studies have shown high levels of interobserver variability for the intermediate (10%-30%) Ki67 LI group [21,26]. The intraobserver variability for the intermediate category (11%-30%) for Ki67 LI was relatively poor according to both the hot spot and average scoring methods, but the average scoring method (ICC, 0.904) for Ki67 LI was better than the hot spot method (ICC, 0.894) [21]. Among 5 pathologists using VA, the correlation was perfect in the low Ki67 LI group (≤10%), whereas it was substantial in the high Ki67 LI group (>30%), and fair to moderate in the intermediate Ki67 LI group (11%-30%) [21]. Another study also showed moderate to high interobserver agreement, especially for the very low and the very high end of the spectrum among 14 raters; however, major disagreements were identified (30%-70%), especially in the mid range of observations [26].
According to the recommendations of the International Ki67 in Breast Cancer Working Group, hot spots should be included in the overall average assessment of Ki67 LI across the whole-tissue section. Furthermore, the group recommends both scoring a minimum of 500 cells for assessing Ki67 LI and evaluating the infiltrative edge of the tumor [9]. In one study, Ki67 assessment in BC showed wide variability among different laboratories (median Ki67 LI ranged from 0.65% to 33.0%; P<.001); this remained significant even when using the same antibody clone (MIB-1, SP6, or 30-9) (17). That study also showed high interlaboratory variability, ranging from 17% to 57% (P<.001) in classifying luminal A–like BCs [17].
The limitations of the current study include the small sample size of our cohort and the random selection of index cases with no exclusion or inclusion criteria.
In conclusion, our study indicates that automated DIA is a better tool for Ki67 assessment in BC than use of VA scoring to eliminate interobserver variability among pathologists.
References
- Denkert C, Budczies J, von Minckwitz G, Wienert S, Loibl S, Klauschen F. Strategies for developing Ki67 as a useful biomarker in breast cancer. Breast. 2015; 24: S67-72.
- Inwald EC, Klinkhammer-Schalke M, Hofstadter F, et al. Ki-67 is a prognostic parameter in breast cancer patients: Results of a large population-based cohort of a cancer registry. Breast Cancer Res Treat. 2013; 139: 539-552.
- Klauschen F, Wienert S, Schmitt WD, et al. Standardized Ki67 Diagnostics Using Automated Scoring-Clinical Validation in the GeparTrio Breast Cancer Study. Clin Cancer Res. 2015; 21: 3651-3657.
- Ryspayeva D, Lyashenko A, Dosenko I, et al. Predictive factors of pathological response to neoadjuvant chemotherapy in patients with breast cancer. J BUON. 2020; 25: 168-175.
- Dowsett M, Smith IE, Ebbs SR, et al. Prognostic value of Ki67 expression after short-term presurgical endocrine therapy for primary breast cancer. J Natl Cancer Inst. 2007; 99: 167-170.
- Goldhirsch A, Wood WC, Coates AS, et al. Strategies for subtypes-dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011; 22: 1736-1747.
- Ellis MJ, Suman VJ, Hoog J, et al. Ki67 Proliferation Index as a Tool for Chemotherapy Decisions During and After Neoadjuvant Aromatase Inhibitor Treatment of Breast Cancer: Results From the American College of Surgeons Oncology Group Z1031 Trial (Alliance). J Clin Oncol. 2017; 35: 1061-1069.
- Robertson J, Dowsett M, Bliss J, et al. Abstract GS1-03: Peri-operative aromatase inhibitor treatment in determining or predicting longterm outcome in early breast cancer – The POETIC* Trial (CRUK/07/015). Cancer Research. 2018; 78: GS1-03-GS1-.
- Dowsett M, Nielsen TO, A’Hern R, et al. Assessment of Ki67 in breast cancer: Recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011; 103: 1656-1664.
- Rimm DL, Leung SCY, McShane LM, et al. An international multicenter study to evaluate reproducibility of automated scoring for assessment of Ki67 in breast cancer. Mod Pathol. 2019; 32: 59-69.
- Kwon AY, Park HY, Hyeon J, et al. Practical approaches to automated digital image analysis of Ki-67 labeling index in 997 breast carcinomas and causes of discordance with visual assessment. PLoS One. 2019; 14: e0212309.
- Zhong F, Bi R, Yu B, Yang F, Yang W, Shui R. A Comparison of Visual Assessment and Automated Digital Image Analysis of Ki67 Labeling Index in Breast Cancer. PLoS One. 2016; 11: e0150505.
- Nassar A, Cohen C, Agersborg SS, et al. A multisite performance study comparing the reading of immunohistochemical slides on a computer monitor with conventional manual microscopy for estrogen and progesterone receptor analysis. Am J Clin Pathol. 2011; 135: 461-467.
- Nassar A, Cohen C, Albitar M, et al. Reading immunohistochemical slides on a computer monitor - A multisite performance study using 180 HER2-stained breast carcinomas. Appl Immunohistochem Mol Morphol. 2011; 19: 212-217.
- Stalhammar G, Robertson S, Wedlund L, et al. Digital image analysis of Ki67 in hot spots is superior to both manual Ki67 and mitotic counts in breast cancer. Histopathology. 2018; 72: 974-989.
- Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986; 1: 307-310.
- Focke CM, Burger H, van Diest PJ, et al. Interlaboratory variability of Ki67 staining in breast cancer. Eur J Cancer. 2017; 84: 219-227.
- Muftah AA, Aleskandarany MA, Al-Kaabi MM, et al. Ki67 expression in invasive breast cancer: The use of tissue microarrays compared with whole tissue sections. Breast Cancer Res Treat. 2017; 164: 341-348.
- Stevanovic L, Choschzick M, Moskovszky L, Varga Z. Variability of predictive markers (hormone receptors, Her2, Ki67) and intrinsic subtypes of breast cancer in four consecutive years 2015-2018. J Cancer Res Clin Oncol. 2019; 145: 2983-2994.
- Bustreo S, Osella-Abate S, Cassoni P, et al. Optimal Ki67 cut-off for luminal breast cancer prognostic evaluation: a large case series study with a long-term follow-up. Breast Cancer Res Treat. 2016; 157: 363-371.
- Shui R, Yu B, Bi R, Yang F, Yang W. An interobserver reproducibility analysis of Ki67 visual assessment in breast cancer. PLoS One. 2015; 10: e0125131.
- Mikami Y, Ueno T, Yoshimura K, et al. Interobserver concordance of Ki67 labeling index in breast cancer: Japan Breast Cancer Research Group Ki67 ring study. Cancer Sci. 2013; 104: 1539-1543.
- Acs B, Pelekanou V, Bai Y, et al. Ki67 reproducibility using digital image analysis: 0041n inter-platform and inter-operator study. Lab Invest. 2019; 99: 107-117.
- Chung YR, Jang MH, Park SY, Gong G, Jung WH, Korean Breast Pathology Ki-67 Study G. Interobserver Variability of Ki-67 Measurement in Breast Cancer. J Pathol Transl Med. 2016; 50: 129-137.
- Romero Q, Bendahl PO, Ferno M, Grabau D, Borgquist S. A novel model for Ki67 assessment in breast cancer. Diagn Pathol. 2014; 9: 118.
- Laenkholm AV, Grabau D, Moller Talman ML, et al. An inter-observer Ki67 reproducibility study applying two different assessment methods: on behalf of the Danish Scientific Committee of Pathology, Danish breast cancer cooperative group (DBCG). Acta Oncol. 2018; 57: 83-89.
- Leung SCY, Nielsen TO, Zabaglo LA, et al. Analytical validation of a standardised scoring protocol for Ki67 immunohistochemistry on breast cancer excision whole sections: An international multicentre collaboration. Histopathology. 2019; 75: 225-235.
- Raap M, Liessem S, Ruschoff J, et al. Quality assurance trials for Ki67 assessment in pathology. Virchows Arch. 2017; 471: 501-508.
MedDocs Publishers
We always work towards offering the best to you. For any queries, please feel free to get in touch with us. Also you may post your valuable feedback after reading our journals, ebooks and after visiting our conferences.